Bootstrapping Confidence Intervals
Source:vignettes/Bootstrapping_Confidence_Intervals.Rmd
Bootstrapping_Confidence_Intervals.Rmd
library(dplyr); library(tidyr); library(purrr); library(ggplot2) # Data wrangling
library(tidyfit) # Model fittingThe combination of .cv = "bootstraps" and
.return_slices = TRUE in tidyfit::regress or
tidyfit::classify makes it very easy to calculate bootstrap
confidence intervals for estimated coefficients. As an additional
convenience function, coef.tidyfit.models includes the
option of adding percentile bootstrap intervals directly. In this short
example, I will calculate and compare bootstrap confidence bands for a
partial least squares regression and a principal components regression
using Boston house price data:
tidyfit handles data scaling internally (i.e. PLSR and
PCR are always fitted on scaled data), however, scaling the data
manually here will give us standardized coefficients, which are easier
to visualize and compare.
Fit the model
Instead of selecting an optimal number of latent components, I define
a preset. This keeps things a little simpler. Note that dropping the
ncomp = 5 argument results the optimal number of components
being selected using bootstrap resampling.
model_frame <- data |>
regress(medv ~ ., m("plsr", ncomp = 5), m("pcr", ncomp = 5),
.cv = "bootstraps", .cv_args = list(times = 100),
.return_slices = TRUE)The coefficients are returned for each slice when
.add_bootstrap_intervals = FALSE (default behavior — see
coef(model_frame)). To obtain bootstrap intervals, I pass
.add_bootstrap_interval = TRUE to coef:
estimates <- coef(model_frame,
.add_bootstrap_interval = TRUE,
.bootstrap_alpha = 0.05)
estimates
#> # A tibble: 28 × 4
#> # Groups: model [2]
#> model term estimate model_info
#> <chr> <chr> <dbl> <list>
#> 1 plsr (Intercept) -0.000809 <tibble [1 × 3]>
#> 2 plsr crim -0.0698 <tibble [1 × 3]>
#> 3 plsr zn 0.0940 <tibble [1 × 3]>
#> 4 plsr indus -0.0271 <tibble [1 × 3]>
#> 5 plsr chas 0.0830 <tibble [1 × 3]>
#> 6 plsr nox -0.202 <tibble [1 × 3]>
#> 7 plsr rm 0.295 <tibble [1 × 3]>
#> 8 plsr age -0.0148 <tibble [1 × 3]>
#> 9 plsr dis -0.333 <tibble [1 × 3]>
#> 10 plsr rad 0.165 <tibble [1 × 3]>
#> # ℹ 18 more rowsThe intervals are nested in model_info:
estimates <- estimates |>
unnest(model_info)
estimates
#> # A tibble: 28 × 6
#> # Groups: model [2]
#> model term estimate ncomp .upper .lower
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 plsr (Intercept) -0.000809 5 0.0418 -0.0377
#> 2 plsr crim -0.0698 5 0.0564 -0.148
#> 3 plsr zn 0.0940 5 0.164 0.0240
#> 4 plsr indus -0.0271 5 0.0466 -0.0963
#> 5 plsr chas 0.0830 5 0.142 0.0186
#> 6 plsr nox -0.202 5 -0.117 -0.302
#> 7 plsr rm 0.295 5 0.413 0.142
#> 8 plsr age -0.0148 5 0.102 -0.107
#> 9 plsr dis -0.333 5 -0.240 -0.432
#> 10 plsr rad 0.165 5 0.233 0.0758
#> # ℹ 18 more rowsPlot the results
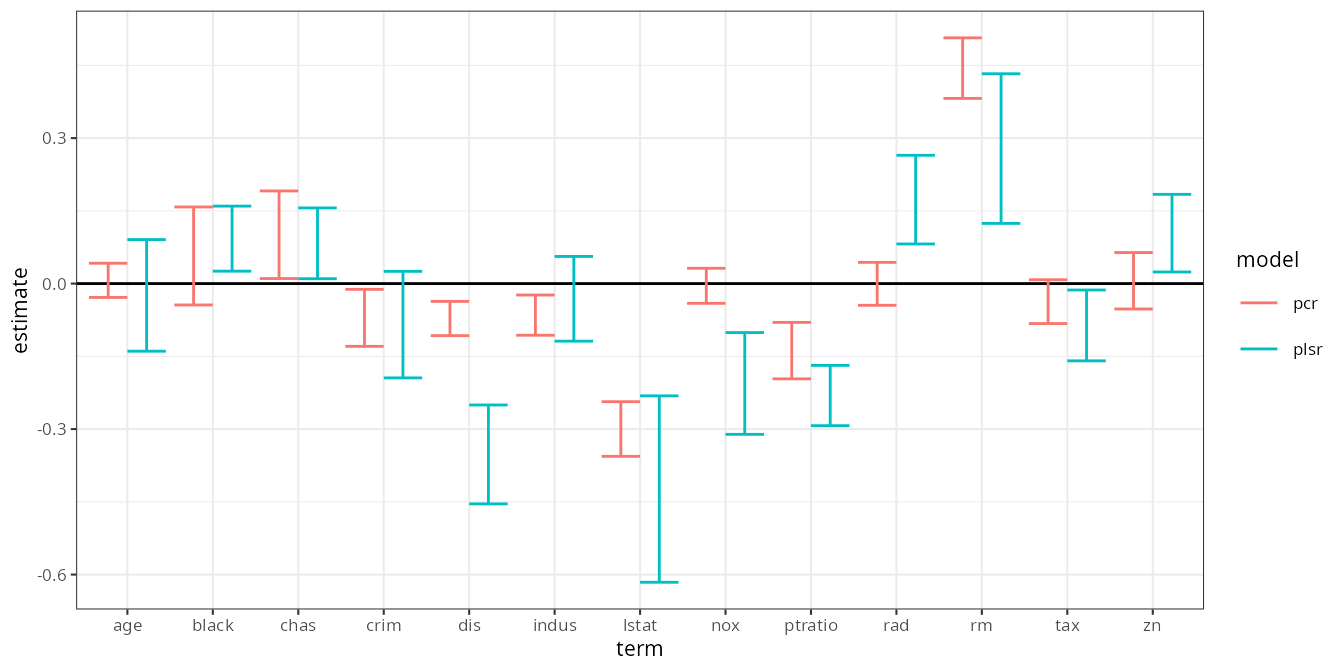
And thus, in a concise workflow, we have 95% bootstrap confidence intervals for the coefficients of a PCR and PLS regression:
estimates |>
filter(term != "(Intercept)") |>
ggplot(aes(term, estimate, color = model)) +
geom_hline(yintercept = 0) +
geom_errorbar(aes(ymin = .lower, ymax = .upper), position = position_dodge()) +
theme_bw(8)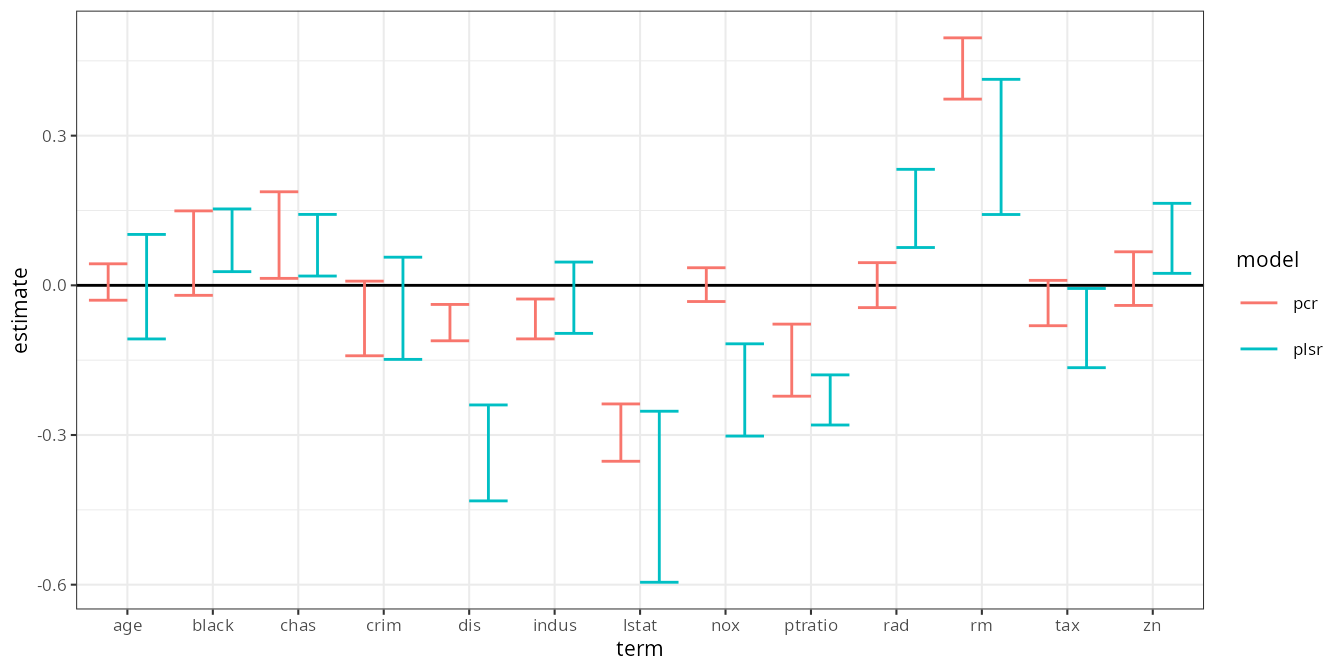
Comparing to built-in jackknife procedure
The pls-package includes built-in functionality to
jackknife confidence intervals for the coefficients. We can compare
these results by passing jackknife = TRUE and
validation = "LOO" to m(), and setting
.cv = "none" (default):
model_frame_jackknife <- data |>
regress(medv ~ ., m("plsr", ncomp = 5, jackknife = TRUE, validation = "LOO"),
m("pcr", ncomp = 5, jackknife = TRUE, validation = "LOO"))
jackknife_estimates <- coef(model_frame_jackknife)Now the coef() generic method also provides standard
errors and
-values
for the coefficients using pls::jack.test:
jackknife_estimates <- jackknife_estimates |>
unnest(model_info) |>
# Create approximate 95% intervals using 2 standard deviations
mutate(.upper = estimate + 2 * std.error, .lower = estimate - 2 * std.error)
jackknife_estimates
#> # A tibble: 28 × 9
#> # Groups: model [2]
#> model term estimate ncomp std.error statistic p.value .upper .lower
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 plsr (Interce… -2.34e-16 5 NA NA NA NA NA
#> 2 plsr crim -8.46e- 2 5 0.0549 -1.54 1.24e- 1 0.0253 -0.194
#> 3 plsr zn 1.04e- 1 5 0.0400 2.60 9.61e- 3 0.184 0.0240
#> 4 plsr indus -3.13e- 2 5 0.0437 -0.718 4.73e- 1 0.0560 -0.119
#> 5 plsr chas 8.30e- 2 5 0.0365 2.28 2.33e- 2 0.156 0.0100
#> 6 plsr nox -2.06e- 1 5 0.0525 -3.92 9.91e- 5 -0.101 -0.311
#> 7 plsr rm 2.78e- 1 5 0.0771 3.61 3.38e- 4 0.433 0.124
#> 8 plsr age -2.43e- 2 5 0.0575 -0.422 6.73e- 1 0.0908 -0.139
#> 9 plsr dis -3.52e- 1 5 0.0510 -6.91 1.47e-11 -0.250 -0.454
#> 10 plsr rad 1.73e- 1 5 0.0457 3.79 1.71e- 4 0.265 0.0817
#> # ℹ 18 more rowsThe plot is almost exactly identical to the bootstrap results above:
jackknife_estimates |>
filter(term != "(Intercept)") |>
ggplot(aes(term, estimate, color = model)) +
geom_hline(yintercept = 0) +
geom_errorbar(aes(ymin = .lower, ymax = .upper), position = position_dodge()) +
theme_bw(8)